M. Nagahashi1, S. Sato2, K. Yuza1, Y. Shimada1, H. Ichikawa1, S. Watanabe3, S. Okuda4, K. Takabe5,6, M. Tsuchida2, T. Wakai1 1Niigata University Graduate School Of Medical And Dental Sciences,Division Of Digestive And General Surgeyr,Niigata City, NIIGATA, Japan 2Niigata University Graduate School Of Medical And Dental Sciences,Division Of Thoracic And Cardiovascular Surgery,Niigata City, NIIGATA, Japan 3Niigata University Graduate School Of Medical And Dental Sciences,Department Of Respiratory Medicine And Infectious Disease,Niigata City, NIIGATA, Japan 4Niigata University Graduate School Of Medical And Dental Sciences,Division Of Bioinformatics,Niigata City, NIIGATA, Japan 5Roswell Park Cancer Institute,Breast Surgery, Department Of Surgical Oncology,Buffalo, NY, USA 6University At Buffalo Jacobs School Of Medicine And Biosciences, The State University Of New York,Department Of Surgery,Buffalo, NY, USA
Introduction:
Although the expectation of immune checkpoint inhibitors, such as anti-PD-1 therapy, is tremendously high based on its dramatic efficacy, only a limited number of patients respond. Even worse, there is no definite biomarker to identify which tumor responds to the therapy. Recent progress in the genomic analysis using next-generation sequencing (NGS) technology has enabled comprehensive detection of mutations and tumor mutation burden (TMB) in cancer patients. The high TMB (TMB-H) tumor is defined as one with high somatic mutational rates, which correlates with the generation of neo-antigens and potential clinical response to immunotherapies. In this study, we analyzed the TMB in patients with lung adenocarcinoma utilizing NGS, and clarified the characteristics of patients with TMB-H in relation to common driver mutations of lung adenocarcinoma and smoking history.
Methods:
Genomic aberrations were determined in Japanese patients with lung adenocarcinoma (N=100) using NGS of 415 known cancer genes, and TMB was determined in each patient. High TMB was defined as 20 or more mutations per megabase of sequenced DNA. Common driver mutations, including ALK, EGFR, ERBB2, ROS, RET, MET, and smoking status were compared with TMB to examine their association.
Results:
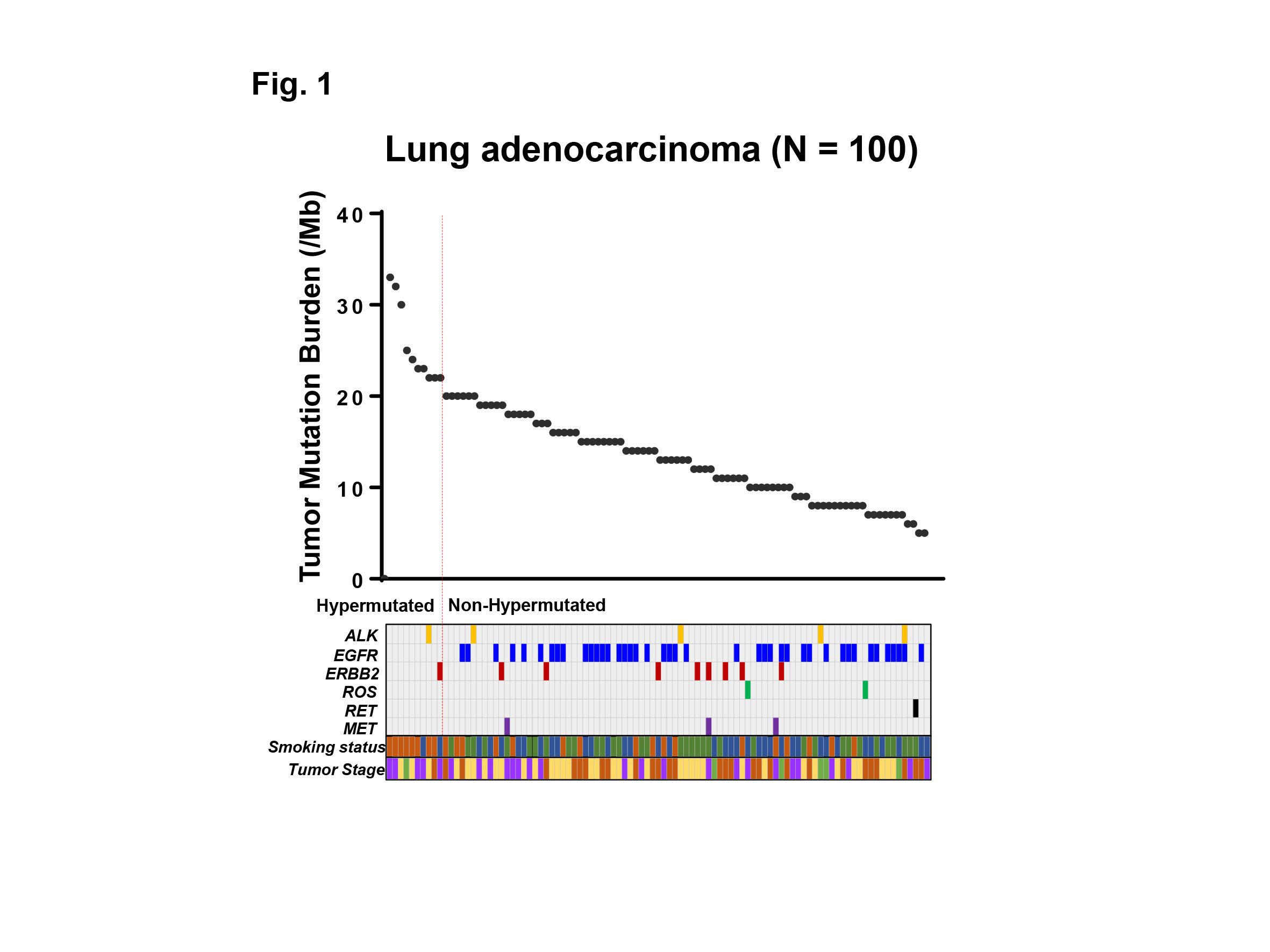
The median TMB was 13.5 (range 5 – 33) per megabase among 100 patients with lung adenocarcinoma examined. Ten out of 100 (10%) patients showed TMB-H, and 90 (90%) patients showed low TMB (TMB-L) (Fig. 1). Only 2 out of 10 (20%) patients with TMB-H had one of common driver mutations (one had ALK, and the other had ERBB2 mutation), while 57 out of 90 (63%) patients with TMB-L had one of the driver mutations including ALK, EGFR, ERBB2, ROS, RET, MET (P<0.05) (Fig. 1). Of note, no EGFR mutation was observed in patients with TMB-H. Eight out of 10 (80%) patients with TMB-H had current smoking history, while 17 out of 90 (19%) patients with TMB-L had the history, respectively (P<0.001). Moreover, a group of current smokers without driver mutations had the highest TMB in our cohort.
Conclusion:
We analyzed the TMB in patients with lung adenocarcinoma utilizing NGS-based analysis. Our data indicates that the common driver mutations and smoking history are associated with TMB-H in lung adenocarcinoma, which may impact treatment strategies for these patients.