J. E. Vidosh1, D. E. Trimble1, M. P. Klueh1, E. Otles1, C. Wu1, L. M. Frydrych1, J. Cuschieri2, M. J. Delano1 1University Of Michigan,Department Of Surgery, Division Of Acute Care Surgery,Ann Arbor, MI, USA 2University Of Washington,Department Of Surgery,Seattle, WA, USA
Introduction: Patients who survive hospitalization following traumatic injury are discharged to either a skilled nursing facility (SNF), rehabilitation center (rehab), or home. Mortality is significantly higher in patients discharged to a SNF compared to rehab or home. The biological mechanisms associated with this increased mortality are unknown; however, the ability to identify patients with an increased mortality risk early during their course is crucial. We hypothesize that distinct genomic expression patterns exist in blood neutrophils, monocytes, and lymphocytes and that these patterns can predict discharge disposition early after injury.
Methods: Inflammation and the Host Response to Injury multicenter database was utilized to review data of 167 blunt trauma patients ≥18 years old. Patient discharge dispositions were identified as SNF, rehab, or home. Patient demographic and clinical data were compared using ANOVA. Affymetrix Glue Grant Human Transcriptome (GG-H) Arrays™ obtained at 7 standardized time points over 28 days were used to assess blood neutrophil, monocyte, and lymphocyte genomic expression. Microarray data was normalized across all subsets using Robust Multi-array Average™ (RMA) software. Pseudo-time ANOVA was incorporated by BRB Array Tools™ version 4.2.1 to compare the genomic changes using a FDR of 0.05%.
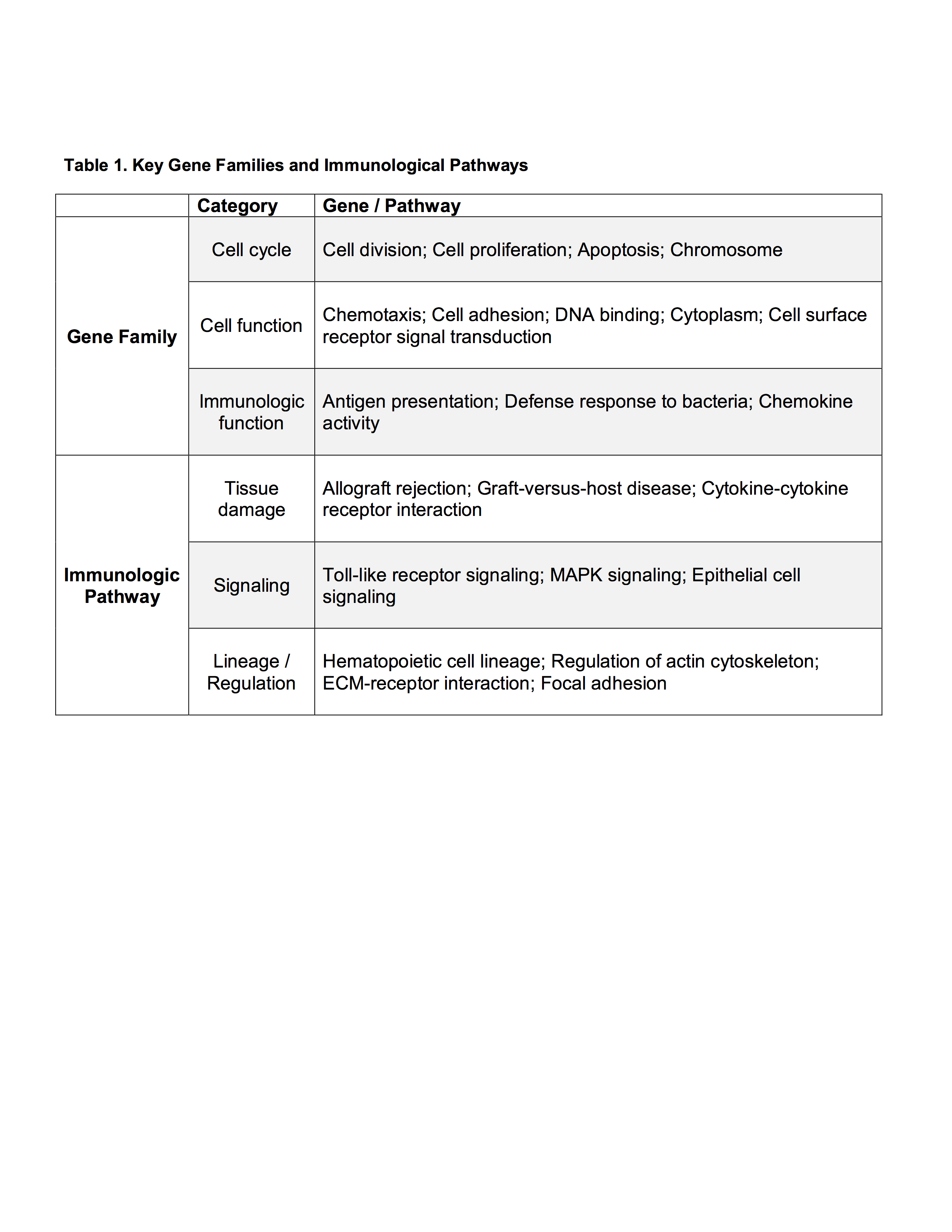
Results: When comparing discharge dispositions, SNF patients experienced greater rates of long-term organ failure (p<0.001). Analysis of genomic expression between discharge groups at 12 hours of admission demonstrated modest differences in lymphocytes and neutrophils, while significant differences were found in monocytes. Monocyte genomic differences persisted over 28 days. At 12 hours, 1187 monocyte genes significantly differed in expression. Over the 28 days, 1058 genes significantly differed. The most important gene families and immunological pathways represented are included in Table 1. The most represented ontological groups are those involved in wound healing and tissue regeneration, which may provoke SNF-associated organ failure.
Conclusion: Monocyte genomic expression differs significantly among severely injured trauma patients depending on discharge disposition, while only modest differences are seen in neutrophil and lymphocyte genomic expression. Monocyte genomic patterns can be detected within 12 hours of admission, persist over time, and are associated with an increased risk of organ failure. Although early genomic expression patterns can identify trauma patients with a poor clinical trajectory requiring SNF placement, more research is necessary to determine if early interventions can alter the clinical course, affect discharge disposition, and improve outcomes.