J. T. George1,2 1Texas A&M University, Department Of Biomedical Engineering, Houston, TX, USA 2School of Engineering Medicine, Houston, TX, USA
Introduction:
Adjuvant immunotherapy has led to durable remission outcomes in many solid cancers. This therapy leverages billions of unique T-cells to confer immunity against foreign epitopes and tumor-associated antigens. Despite improvements in patient outcomes using T-cell immunotherapies, complete remission is limited by cancer immune evasion. Prior efforts have quantified the effect of immune system function of cancer control and immunotherapeutic efficacy[1,2]. We introduce a framework for quantifying the interaction between an adaptive immune system recognizing tumor-associated antigens (TAAs) on the surface of dominant cancer clones. We investigate the resulting evolutionary dynamics assuming tumors either passively evade recognition or actively escape adaptive immune detection. Using this framework, we represent cancer evasion on a spectrum of passive to active evasion. Our model serves as a first attempt at characterizing the extent of optimality of cancer evolution in the presence of the adaptive immune system.
Methods: We apply large-scale in-silico numerical simulations and discrete-time stochastic process theory to track the time dynamics of the number of detectable TAAs. Relevant modeling features include immune recognition and cancer evasion rates, and TAA availability, antigenicity, and accumulation rate. This model quantifies the likelihood and escape time of cancer immune evasion. Analytical equations are compared to largescale numerical simulations via a modified Gillespie algorithm to quantify model accuracy.
Results:
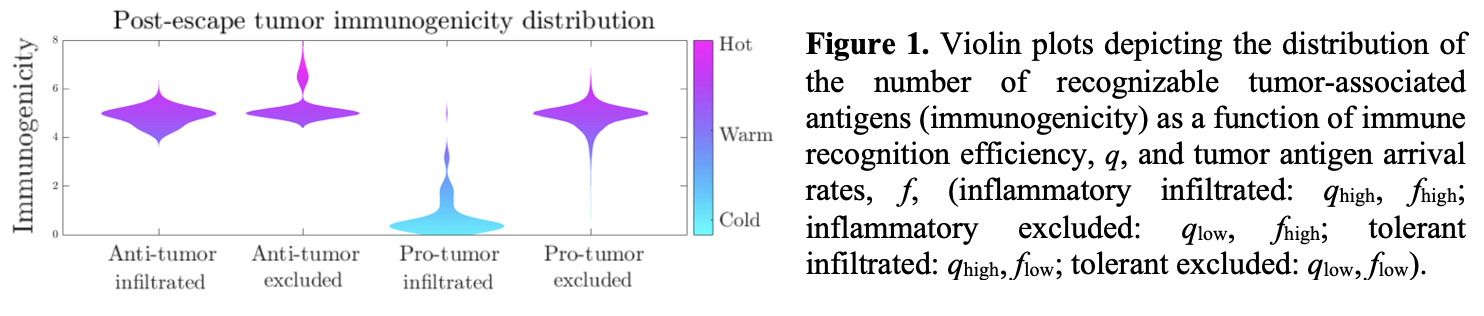
We correlates aggressiveness significantly with disease incidence across a variety of temporally varying recognition policies. Moreover, we find that the distribution of aggressive cancer immunogenicity following immune escape depends on the recognition efficiency of the adaptive immune system and the average arrival rate of new TAAs (Fig. 1). Substantial variety in evolutionary trajectories agree with prior clinical observations and together explain effects of the tumor-immune microenvironment on the generation of immunogenically hot or cold tumors.
Conclusion:
The development of a computational framework to track T-cell recognition (and cancer generation) of TAAs enables analysis of the likelihood and timing of immune escape, in addition to the predicted distribution of post-escape antigens. When applied to a variety of cancer microenvironments, our model can predict the clinically observed diversity in post-escape immunogenicity, which impacts the predicted efficacy of immune detection.
Acknowledgments:
JTG is a CPRIT Scholar in Cancer Research and is supported by CPRIT grant (RR210080).
[1] George JT, Levine H. Can Res 2020;80:811-819.
[2] George JT, Levine H. J Theor Biol 2018;458:148-155.