M. Mirza1, J. Baechle1, P. Marincola1, J. Vaz2, A. Naveed1, D. Hanna1, M. K. Jolly2, K. Idrees1 1Vanderbilt University Medical Center, Division Of Surgical Oncology, Department Of Surgery, Vanderbilt-Ingram Cancer Center, Nashville, TN, USA 2Indian Institute of Science, Centre For BioSystems Science And Engineering, Banglore, BANGLORE, India
Introduction: Colorectal peritoneal metastases (CPM) portend worse clinical outcomes in comparison to hematogenous metastasis (HM). Yet the CPM disease biology remains poorly characterized. Herein, we aim to elucidate the role of collagen remodeling in CPM as a molecular pathway driving cancer progression and its interaction with TAMs in the tumor microenvironment (TME).
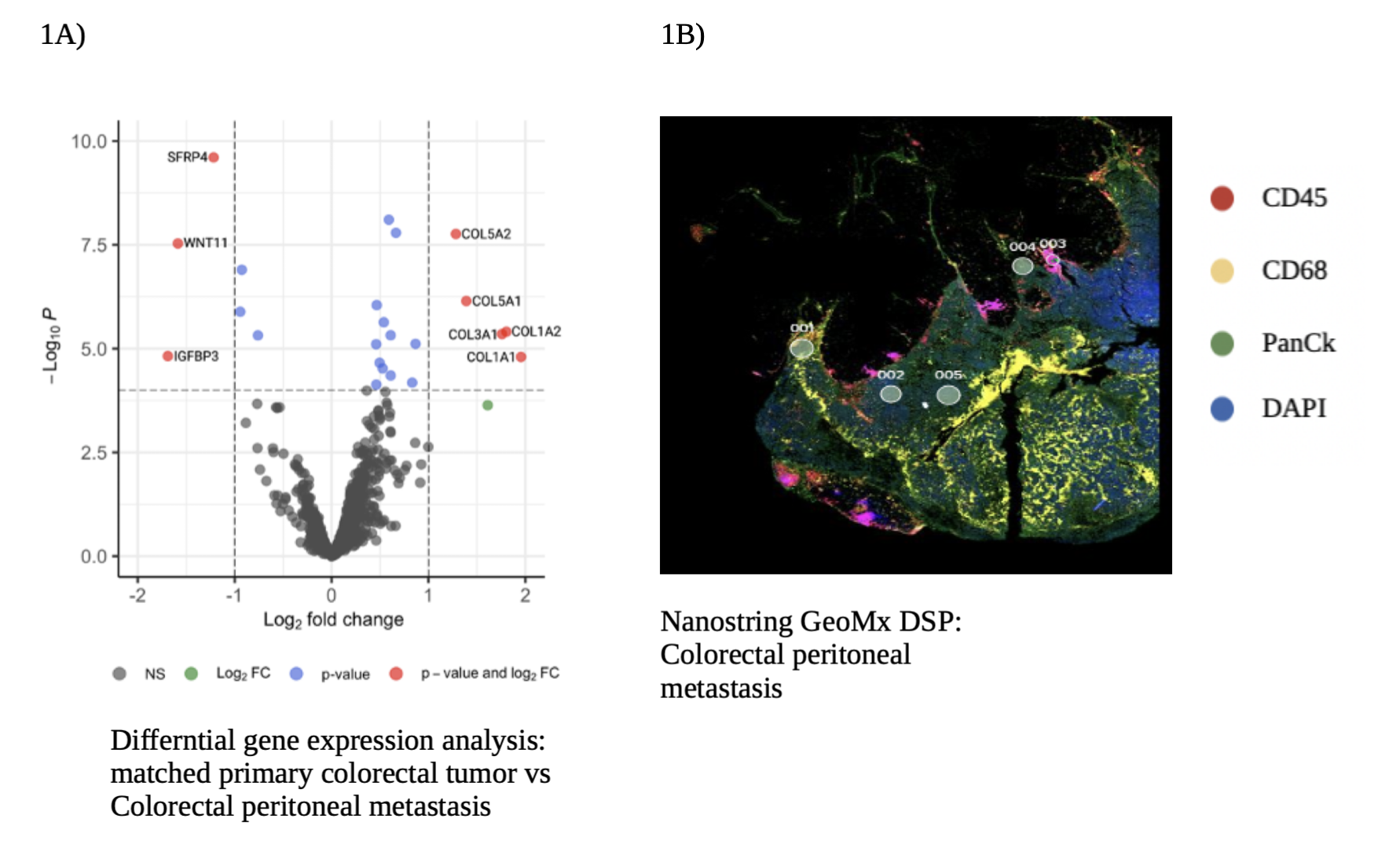
Methods: 70 matched cancer samples, surgically resected from 17 patients with colorectal metastasis were analyzed with bulk RNA-sequencing. Nanostring GeoMx digital spatial profiler (DSP) platform was used to for spatial localization and transcriptomics of 8 matched samples from 2 patients with colorectal metastasis. Gene Set Enrichment Analysis (GSEA) was used to calculate relative expression of collagen gene signature and the Ingenuity Pathway Analysis (IPA) was used for differential pathway analysis.
Results: All samples included in the bulk RNA-sequencing analysis were categorized into four groups. Primary tumors (n=21), HM which included liver and lung (n=21), lymph node metastasis (n=10) and CPM (n=18). Collagen gene signature was statistically significant in the CPM when compared to primary tumors and HM (p = 0.008), revealing extensive collagen remodeling in CPM. Transcriptomic analysis on Nanostring GeoMx DSP showed similar results, with a differential expression for COL5A2, COL5A1, COL3A1, COL1A2, COL1A1 genes in CPM when compared to matched primary tumors (Figure 1A). IPA showed an upregulation of extracellular structure organization and collagen fibrils organization pathways in CPM. Spatial visualization of the TME revealed an infiltration of TAMs in CPM, shown as abundant immunofluorescence staining for CD68, a surface protein highly expressed by macrophages (Figure 1B). mRNA expression analysis of CPM showed a preponderance of pro-tumorigenic M2 TAMs, compared to anti-tumorigenic M1 TAMs. Further analysis of M2 TAMs in CPM, showed a significant activation of TGF-β signaling, known to play a key role in extra-cellular remodeling.
Conclusion: CPM are characterized by widespread collagen remodeling and infiltration of tumor associated macrophages. Our study indicates that collagen remodeling in CPM, may be driven by TAMs. More mechanistic studies will be required to validate our results.